Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Drop outliers and interpolate#
Filter out points with low confidence scores and interpolate over missing values.
Imports#
import json
from movement import sample_data
from movement.filtering import filter_by_confidence, interpolate_over_time
from movement.kinematics import compute_velocity
Load a sample dataset#
ds = sample_data.fetch_dataset("DLC_single-wasp.predictions.h5")
print(ds)
<xarray.Dataset> Size: 61kB
Dimensions: (time: 1085, space: 2, keypoints: 2, individuals: 1)
Coordinates:
* time (time) float64 9kB 0.0 0.025 0.05 0.075 ... 27.05 27.07 27.1
* space (space) <U1 8B 'x' 'y'
* keypoints (keypoints) <U7 56B 'head' 'stinger'
* individuals (individuals) <U12 48B 'individual_0'
Data variables:
position (time, space, keypoints, individuals) float64 35kB 1.086e+03...
confidence (time, keypoints, individuals) float64 17kB 0.05305 ... 0.0
Attributes:
source_software: DeepLabCut
ds_type: poses
fps: 40.0
time_unit: seconds
source_file: /home/runner/.movement/data/poses/DLC_single-wasp.predi...
frame_path: /home/runner/.movement/data/frames/single-wasp_frame-10...
We see that the dataset contains the 2D pose tracks and confidence scores for a single wasp, generated with DeepLabCut. The wasp is tracked at two keypoints: “head” and “stinger” in a video that was recorded at 40 fps and lasts for approximately 27 seconds.
Visualise the pose tracks#
Since the data contains only a single wasp, we use
xarray.DataArray.squeeze() to remove
the dimension of length 1 from the data (the individuals dimension).
ds.position.squeeze().plot.line(
x="time", row="keypoints", hue="space", aspect=2, size=2.5
)
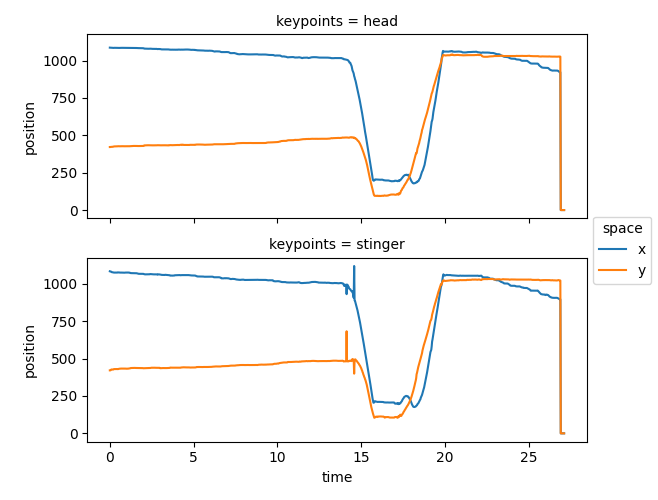
<xarray.plot.facetgrid.FacetGrid object at 0x7fc0c95336b0>
We can see that the pose tracks contain some implausible “jumps”, such as the big shift in the final second, and the “spikes” of the stinger near the 14th second. Perhaps we can get rid of those based on the model’s reported confidence scores?
Visualise confidence scores#
The confidence scores are stored in the confidence data variable.
Since the predicted poses in this example have been generated by DeepLabCut,
the confidence scores should be likelihood values between 0 and 1.
That said, confidence scores are not standardised across pose
estimation frameworks, and their ranges can vary. Therefore,
it’s always a good idea to inspect the actual confidence values in the data.
Let’s first look at a histogram of the confidence scores. As before, we use
xarray.DataArray.squeeze() to remove the individuals dimension
from the data.
ds.confidence.squeeze().plot.hist(bins=20)
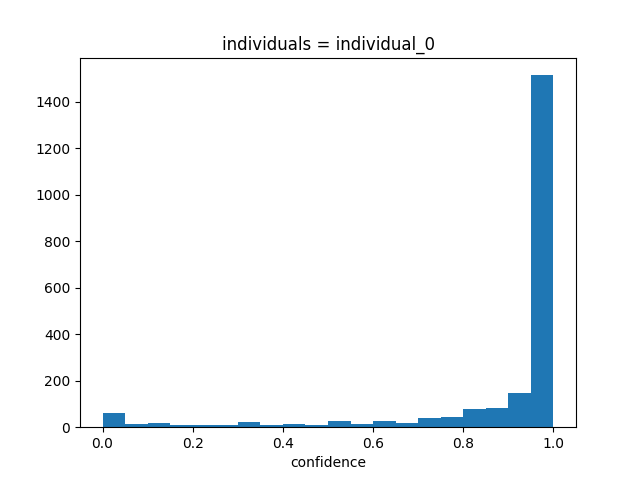
(array([ 61., 13., 16., 10., 10., 8., 21., 11., 14.,
11., 26., 13., 28., 19., 39., 44., 79., 84.,
149., 1514.]), array([0. , 0.04999823, 0.09999646, 0.14999469, 0.19999292,
0.24999115, 0.29998938, 0.34998761, 0.39998584, 0.44998407,
0.4999823 , 0.54998053, 0.59997876, 0.64997699, 0.69997522,
0.74997345, 0.79997168, 0.84996991, 0.89996814, 0.94996637,
0.99996459]), <BarContainer object of 20 artists>)
Based on the above histogram, we can confirm that the confidence scores indeed range between 0 and 1, with most values closer to 1. Now let’s see how they evolve over time.
ds.confidence.squeeze().plot.line(
x="time", row="keypoints", aspect=2, size=2.5
)
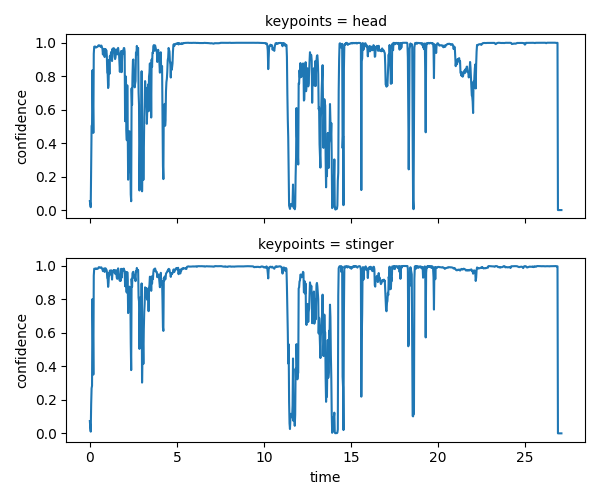
<xarray.plot.facetgrid.FacetGrid object at 0x7fc0c96abf80>
Encouragingly, some of the drops in confidence scores do seem to correspond
to the implausible jumps and spikes we had seen in the position.
We can use that to our advantage, by leveraging functions from the
movement.filtering module.
Filter out points with low confidence#
Using movement.filtering.filter_by_confidence(),
we can filter out points with confidence scores below a certain threshold.
This function takes position and confidence as required arguments,
and accepts an optional threshold parameter,
which defaults to threshold=0.6 unless specified otherwise.
Setting print_report=True, will make the function print a report
on the number of NaN values before and after the filtering operation.
This is False by default, but can be useful for understanding
and debugging the impact of filtering on the data.
We will use xarray.Dataset.update() to update ds in-place
with the filtered position.
ds.update(
{
"position": filter_by_confidence(
ds.position, ds.confidence, print_report=True
)
}
)
No missing points (marked as NaN) in input.
Missing points (marked as NaN) in output:
keypoints head stinger
individuals
individual_0 121/1085 (11.15%) 93/1085 (8.57%)
We can see that the filtering operation has introduced NaN values in the
position data variable. Let’s visualise the filtered data.
ds.position.squeeze().plot.line(
x="time", row="keypoints", hue="space", aspect=2, size=2.5
)
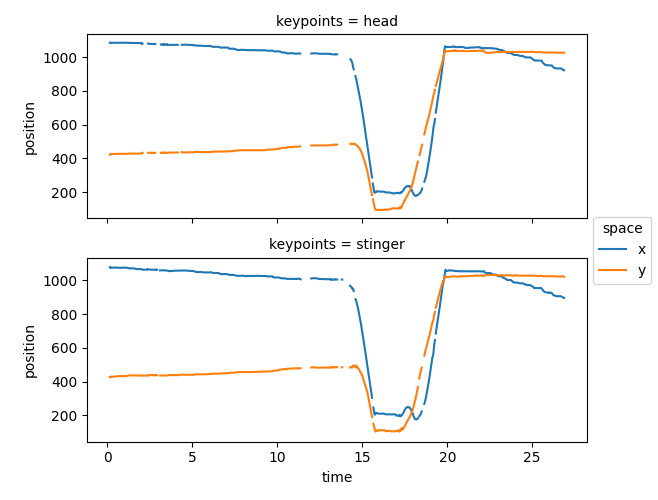
<xarray.plot.facetgrid.FacetGrid object at 0x7fc0c96ab4d0>
Here we can see that gaps (consecutive NaNs) have appeared in the pose tracks, some of which are over the implausible jumps and spikes we had seen earlier. Moreover, most gaps seem to be brief, lasting < 1 second (or 40 frames).
Interpolate over missing values#
Using movement.filtering.interpolate_over_time(), we can
interpolate over gaps we’ve introduced in the pose tracks.
Here we use the default linear interpolation method (method="linear")
and interpolate over gaps of 40 frames or less (max_gap=40).
The default max_gap=None would interpolate over all gaps, regardless of
their length, but this should be used with caution as it can introduce
spurious data. The print_report argument acts as described above.
ds.update(
{
"position": interpolate_over_time(
ds.position, max_gap=40, print_report=True
)
}
)
Missing points (marked as NaN) in input:
keypoints head stinger
individuals
individual_0 121/1085 (11.15%) 93/1085 (8.57%)
Missing points (marked as NaN) in output:
keypoints head stinger
individuals
individual_0 15/1085 (1.38%) 15/1085 (1.38%)
We see that most, but not all, NaN values have disappeared, meaning that most gaps were indeed shorter than 40 frames. Let’s visualise the interpolated pose tracks.
ds.position.squeeze().plot.line(
x="time", row="keypoints", hue="space", aspect=2, size=2.5
)
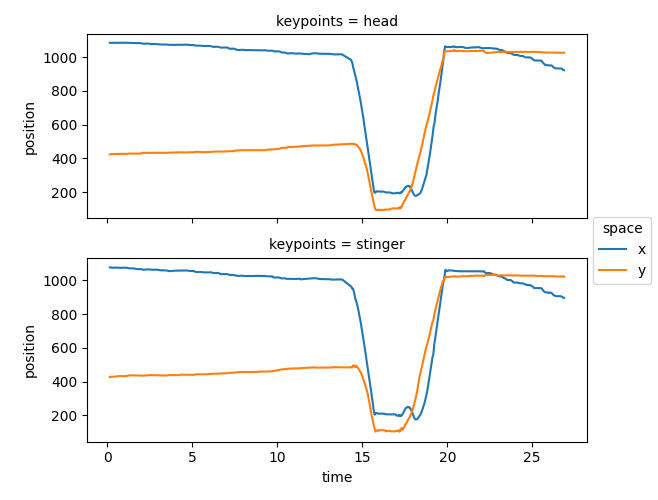
<xarray.plot.facetgrid.FacetGrid object at 0x7fc0ba6832c0>
Note
By default, interpolation does not apply to gaps at either end of the time
series. In our case, the last few frames removed by filtering were not
replaced by interpolation. Passing fill_value="extrapolate" to the
interpolate_over_time()
function would extrapolate the data to also fill the gaps at either end.
In general, you may pass any keyword arguments that are acceptable as
**kwargs by xarray.DataArray.interpolate_na(), which in turn
passes them to its underlying interpolation methods.
Log of processing steps#
So, far we’ve processed the pose tracks first by filtering out points with
low confidence scores, and then by interpolating over missing values.
The order of these operations and the parameters with which they were
performed are saved in the log attribute of the position data array.
This is useful for keeping track of the processing steps that have been
applied to the data. Let’s inspect the log:
print(ds.position.log)
[
{
"operation": "filter_by_confidence",
"datetime": "2025-09-19 14:26:56.492915",
"confidence": "<xarray.DataArray 'confidence' (time: 1085, keypoints: 2, individuals: 1)> Size: 17kB\n0.05305 0.07366 0.03532 0.03293 0.01707 0.01022 ... 0.0 0.0 0.0 0.0 0.0 0.0\nCoordinates:\n * time (time) float64 9kB 0.0 0.025 0.05 0.075 ... 27.05 27.07 27.1\n * keypoints (keypoints) <U7 56B 'head' 'stinger'\n * individuals (individuals) <U12 48B 'individual_0'",
"threshold": "0.6",
"print_report": "True"
},
{
"operation": "interpolate_over_time",
"datetime": "2025-09-19 14:26:57.654830",
"method": "'linear'",
"max_gap": "40",
"print_report": "True"
}
]
If you want to retrieve a certain parameter from the log, you can use
json.loads() to parse the log string into a list of dictionaries.
log_entries = json.loads(ds.position.log) # A list of dictionaries
# Print the 'max_gap' parameter from the last log entry
print(
log_entries[-1].get("max_gap", None) # None if "max_gap" is not present
)
40
Filtering multiple data variables#
We can also apply the same filtering operation to
multiple data variables in ds at the same time.
For instance, to filter both position and velocity data variables
in ds, based on the confidence scores, we can specify a dictionary
with the data variable names as keys and the corresponding filtered
DataArrays as values. Then we can once again use
xarray.Dataset.update() to update ds in-place
with the filtered data variables.
# Add velocity data variable to the dataset
ds["velocity"] = compute_velocity(ds.position)
# Create a dictionary mapping data variable names to filtered DataArrays
# We disable report printing for brevity
update_dict = {
var: filter_by_confidence(ds[var], ds.confidence)
for var in ["position", "velocity"]
}
# Use the dictionary to update the dataset in-place
ds.update(update_dict)
Total running time of the script: (0 minutes 1.936 seconds)