Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Load and upsample bounding box tracks#
Load bounding box tracks and upsample them to match the video frame rate.
Imports#
# For interactive plots: install ipympl with `pip install ipympl` and uncomment
# the following line in your notebook
# %matplotlib widget
import csv
import math
import os
import sleap_io as sio
from matplotlib import pyplot as plt
from movement import sample_data
from movement.filtering import interpolate_over_time
from movement.io import load_bboxes, save_bboxes
Load sample dataset#
In this tutorial, we will use a sample bounding boxes dataset with a single individual (a crab). The clip is part of the Moving Camouflaged Animals Dataset (MoCA) dataset.
We will also download the associated video for visualising the data later.
dataset_dict = sample_data.fetch_dataset_paths(
"VIA_single-crab_MOCA-crab-1.csv",
with_video=True, # download associated video
)
file_path = dataset_dict["bboxes"]
print(file_path)
ds = load_bboxes.from_via_tracks_file(
file_path, use_frame_numbers_from_file=True
)
/home/runner/.movement/data/bboxes/VIA_single-crab_MOCA-crab-1.csv
The loaded dataset is made up of three data arrays:
position, shape, and confidence.
print(ds)
<xarray.Dataset> Size: 2kB
Dimensions: (time: 35, space: 2, individuals: 1)
Coordinates:
* time (time) int64 280B 0 5 10 15 20 25 ... 145 150 155 160 165 167
* space (space) <U1 8B 'x' 'y'
* individuals (individuals) <U4 16B 'id_1'
Data variables:
position (time, space, individuals) float64 560B 1.118e+03 ... 401.9
shape (time, space, individuals) float64 560B 320.1 153.2 ... 120.1
confidence (time, individuals) float64 280B nan nan nan ... nan nan nan
Attributes:
source_software: VIA-tracks
ds_type: bboxes
time_unit: frames
source_file: /home/runner/.movement/data/bboxes/VIA_single-crab_MOCA...
We can see that the coordinates in the time dimension are expressed in frames, and that we only have data for 1 in 5 frames of the video, plus the last frame (frame number 167).
print(ds.time)
<xarray.DataArray 'time' (time: 35)> Size: 280B
0 5 10 15 20 25 30 35 40 45 50 ... 120 125 130 135 140 145 150 155 160 165 167
Coordinates:
* time (time) int64 280B 0 5 10 15 20 25 30 ... 145 150 155 160 165 167
In the following sections of the notebook we will explore options to upsample the dataset by filling in values for the video frames with no data.
Inspect associated video#
The video associated to the data contains all 168 frames.
video_path = dataset_dict["video"]
video = sio.load_video(video_path)
n_frames, height, width, channels = video.shape
print(f"Number of frames: {n_frames}")
print(f"Frame size: {width}x{height}")
print(f"Number of channels: {channels}")
Number of frames: 168
Frame size: 1280x720
Number of channels: 3
Let’s inspect the first 6 frames of the video for which we have annotations, and plot the annotated bounding box and centroid at each frame.
# set last frame to plot
end_frame_idx = 25
# create list of frames to loop over with step=5
list_frames = list(range(0, end_frame_idx + 1, 5))
# initialise figure
fig = plt.figure(figsize=(8, 8)) # width, height
# loop over selected frames and plot the data
for i, frame_idx in enumerate(list_frames):
# add subplot axes
ax = plt.subplot(math.ceil(len(list_frames) / 2), 2, i + 1)
# plot frame
ax.imshow(video[frame_idx])
# plot box at this frame
top_left_corner = (
ds.position.sel(time=frame_idx).data
- ds.shape.sel(time=frame_idx).data / 2
).squeeze()
bbox = plt.Rectangle(
xy=tuple(top_left_corner),
width=ds.shape.sel(time=frame_idx, space="x").item(),
height=ds.shape.sel(time=frame_idx, space="y").item(),
edgecolor="red",
facecolor="none",
linewidth=1.5,
label="current frame",
)
ax.add_patch(bbox)
# plot box's centroid at this frame with red ring
ax.scatter(
x=ds.position.sel(time=frame_idx, space="x"),
y=ds.position.sel(time=frame_idx, space="y"),
s=15,
color="red",
)
# plot past centroid positions in blue
if frame_idx > 0:
ax.scatter(
x=ds.position.sel(time=slice(0, frame_idx - 1), space="x"),
y=ds.position.sel(time=slice(0, frame_idx - 1), space="y"),
s=10,
color="tab:blue",
label="past frames",
)
# plot future centroid positions in white
ax.scatter(
x=ds.position.sel(time=slice(frame_idx + 1, end_frame_idx), space="x"),
y=ds.position.sel(time=slice(frame_idx + 1, end_frame_idx), space="y"),
s=10,
color="white",
label="future frames",
)
# set title and labels
ax.set_title(f"Frame {frame_idx}")
ax.set_xlabel("x (pixles)")
ax.set_ylabel("y (pixels)")
ax.set_xlabel("")
if frame_idx == 1:
ax.legend()
fig.tight_layout()
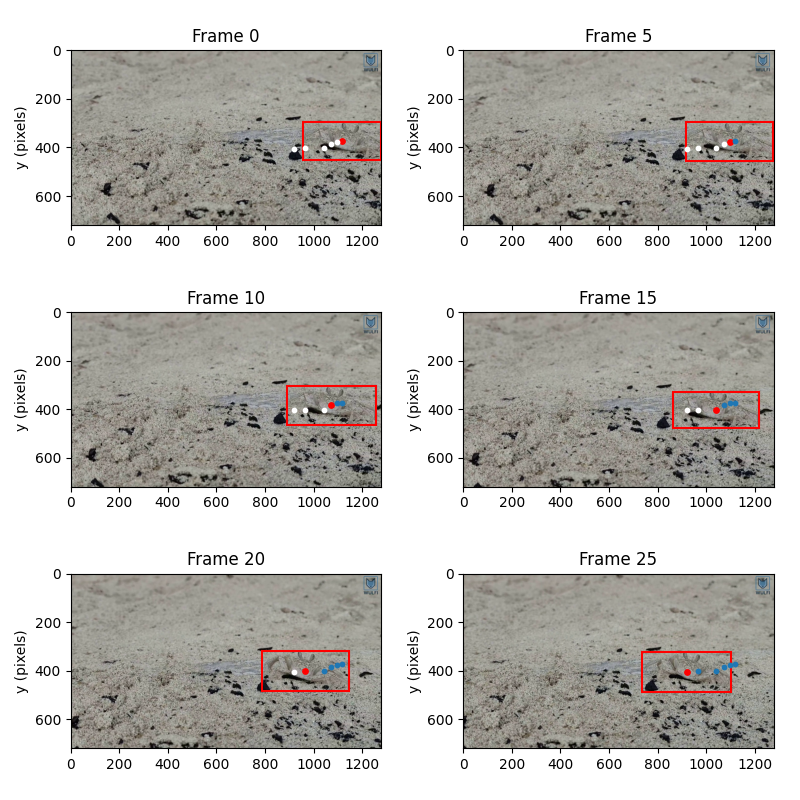
We used xarray.DataArray.sel() to select the data for the
relevant frames directly.
The centroid at each frame is marked with a red marker. The past centroid positions are shown in blue and the future centroid positions in white. Note that in this case the camera is not static relative to the environment.
Fill in empty values with forward filling#
We can fill in the frames with empty values for the position and
shape arrays by taking the last valid value in time. In this way, a
box’s position and shape stay constant if for a current frame the box
has no annotation defined.
ds_ff = ds.reindex(
{"time": list(range(ds.time[-1].item() + 1))},
method="ffill", # propagate last valid index value forward
)
We can verify with a plot that the empty values have been filled in using the last valid value in time.
For this we define a convenience function to plot the x and y coordinates
of the position and shape arrays, for the input dataset and for
a filled one.
def plot_position_and_shape_xy_coords(ds_input_data, ds_filled, color_filled):
"""Compare the x and y coordinates of the position and shape arrays in time
for the input and filled datasets.
"""
fig, axs = plt.subplots(2, 2, figsize=(8, 6))
for row in range(axs.shape[0]):
space_coord = ["x", "y"][row]
for col in range(axs.shape[1]):
ax = axs[row, col]
data_array_str = ["position", "shape"][col]
# plot original data
ax.scatter(
x=ds_input_data.time,
y=ds_input_data[data_array_str].sel(
individuals="id_1", space=space_coord
),
marker="o",
color="black",
label="original data",
)
# plot forward filled data
ax.plot(
ds_filled.time,
ds_filled[data_array_str].sel(
individuals="id_1", space=space_coord
),
marker=".",
linewidth=1,
color=color_filled,
label="upsampled data",
)
# set axes labels and legend
ax.set_ylabel(f"{space_coord} (pixels)")
if row == 0:
ax.set_title(f"Bounding box {data_array_str}")
if col == 1:
ax.legend()
if row == 1:
ax.set_xlabel("time (frames)")
fig.tight_layout()
In the plot below, the original position and shape data is shown
in black, while the forward-filled values are shown in green.
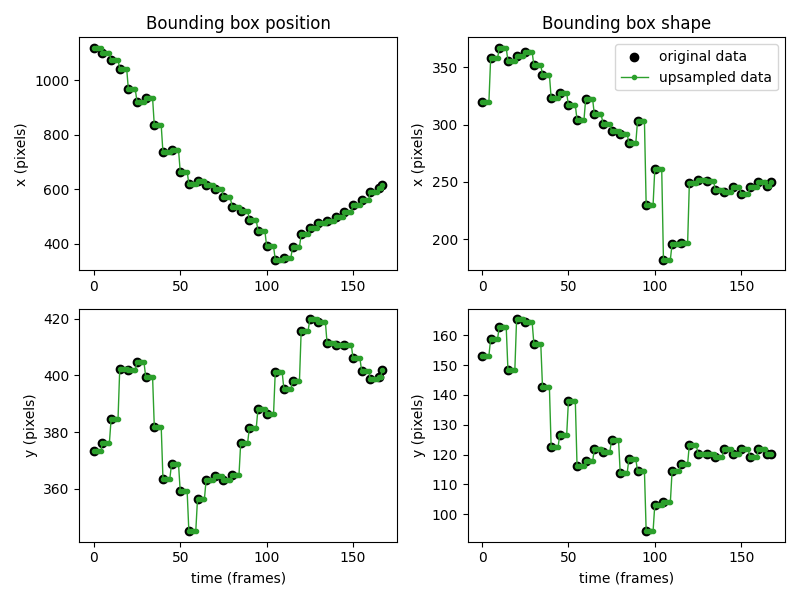
Fill in empty values with NaN#
Alternatively, we can fill in the empty frames with NaN values. This can be useful if we want to interpolate later.
ds_nan = ds.reindex(
{"time": list(range(ds.time[-1].item() + 1))},
method=None, # default
)
Like before, we can verify with a plot that the empty values have been filled with NaN values.
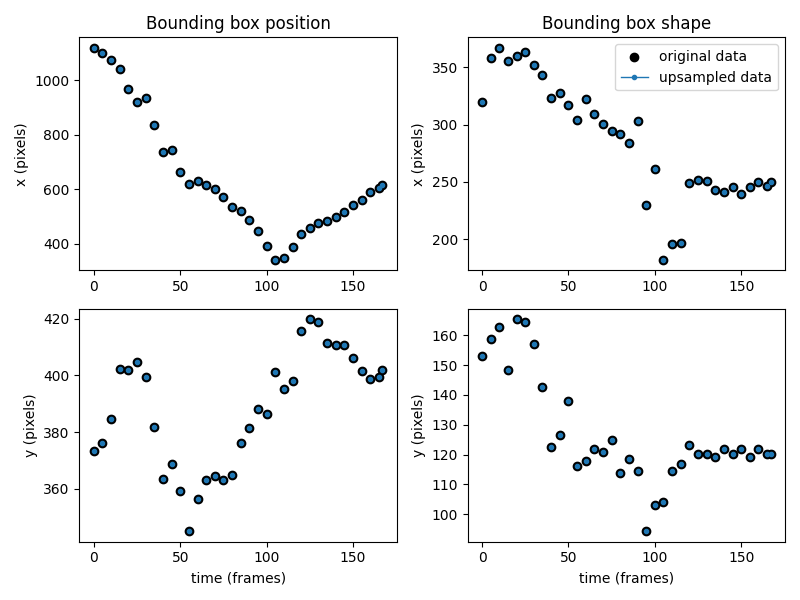
We can further confirm we have NaNs where expected by printing the first few frames of the data.
print("Position data array (first 10 frames):")
print(ds_nan.position.isel(time=slice(0, 10), individuals=0).data)
print("----")
print("Shape data array (first 10 frames):")
print(ds_nan.shape.isel(time=slice(0, 10), individuals=0).data)
Position data array (first 10 frames):
[[1117.895 373.3035]
[ nan nan]
[ nan nan]
[ nan nan]
[ nan nan]
[1098.141 376.1255]
[ nan nan]
[ nan nan]
[ nan nan]
[ nan nan]]
----
Shape data array (first 10 frames):
[[320.09 153.191]
[ nan nan]
[ nan nan]
[ nan nan]
[ nan nan]
[357.984 158.835]
[ nan nan]
[ nan nan]
[ nan nan]
[ nan nan]]
Linearly interpolate NaN values#
We can instead fill in the empty values in the dataset by linearly
interpolating the position and shape data arrays. In this way,
we would be assuming that the centroid of the bounding box moves linearly
between the two annotated values, and its width and height change linearly
as well.
We use the dataset with NaN values as an input to
movement.filtering.interpolate_over_time().
ds_interp = ds_nan.copy()
for data_array_str in ["position", "shape"]:
ds_interp[data_array_str] = interpolate_over_time(
data=ds_interp[data_array_str],
method="linear",
max_gap=None,
print_report=False,
)
Like before, we can visually check that the empty data has been imputed as
expected by plotting the x and y coordinates of the position
and shape arrays
in time.
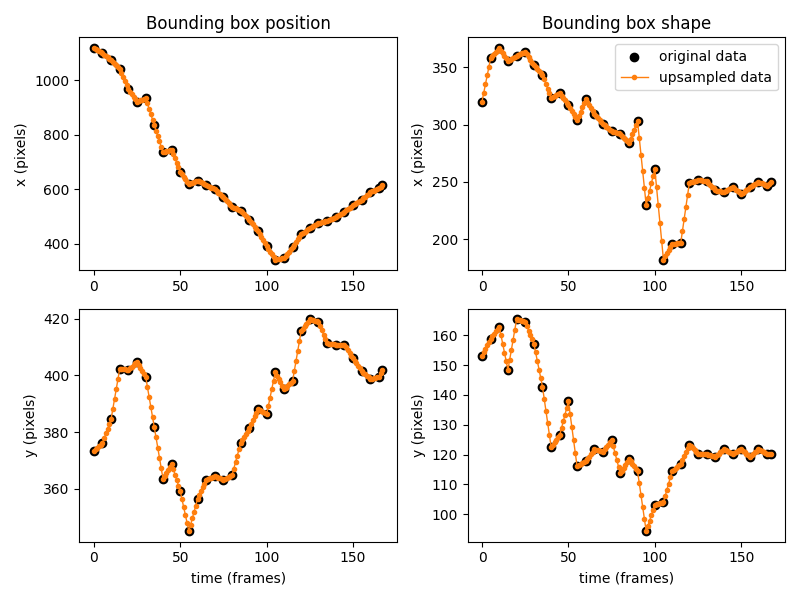
The plot above shows that between the original data points (in black), the data is assumed to evolve linearly (in orange).
Compare methods#
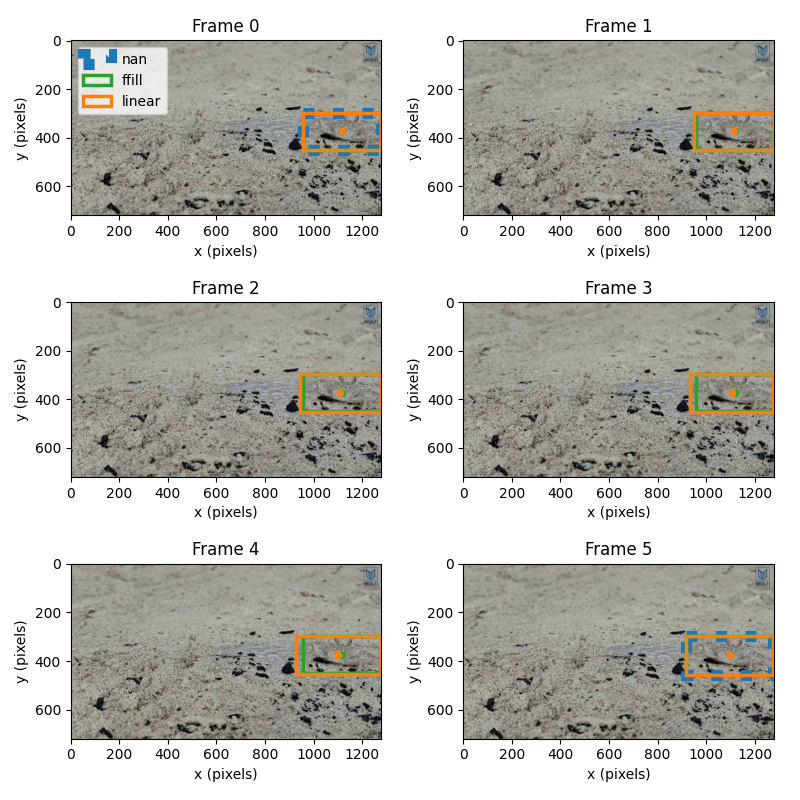
We can now qualitatively compare the bounding boxes computed with the three different filling methods we have seen: forward filling, NaN filling and linear interpolation
In the plot below, the NaN-filled data are plotted in blue, the forward filled values are plotted in orange, and the linearly interpolated values are shown in green.
# initialise figure
fig = plt.figure(figsize=(8, 8))
list_colors = ["tab:blue", "tab:green", "tab:orange"]
# loop over frames
for frame_idx in range(6):
# add subplot axes
ax = plt.subplot(3, 2, frame_idx + 1)
# plot frame
ax.imshow(video[frame_idx])
# plot bounding box for each dataset
for ds_i, ds_filled in enumerate([ds_nan, ds_ff, ds_interp]):
# plot box
top_left_corner = (
ds_filled.position.sel(time=frame_idx).data
- ds_filled.shape.sel(time=frame_idx).data / 2
).squeeze()
bbox = plt.Rectangle(
xy=tuple(top_left_corner),
width=ds_filled.shape.sel(time=frame_idx, space="x").item(),
height=ds_filled.shape.sel(time=frame_idx, space="y").item(),
edgecolor=list_colors[ds_i],
facecolor="none",
# make line for NaN dataset thicker and dotted
label=["nan", "ffill", "linear"][ds_i],
linewidth=[8, 2.5, 2.5][ds_i],
linestyle=["dotted", "solid", "solid"][ds_i],
)
ax.add_patch(bbox)
# plot centroid
ax.scatter(
x=ds_filled.position.sel(time=frame_idx, space="x"),
y=ds_filled.position.sel(time=frame_idx, space="y"),
s=20,
color=list_colors[ds_i],
)
# set title and labels
ax.set_title(f"Frame {frame_idx}")
ax.set_xlabel("x (pixels)")
ax.set_ylabel("y (pixels)")
if frame_idx == 0:
ax.legend()
fig.tight_layout()
Export as a VIA tracks .csv file#
Let’s assume the dataset with the forward filled values is the best suited
for our task. We can now export the computed values to a VIA tracks .csv
file that is loadable in the movement napari widget
and in the VGG Image Annotator (VIA)
software.
via_tracks_filepath = "tracking_output_via_tracks.csv"
save_bboxes.to_via_tracks_file(ds_ff, via_tracks_filepath)
PosixPath('tracking_output_via_tracks.csv')
Alternatively, we can save the bounding boxes’ trajectories to a
.csv file with a custom header using the standard Python library
csv.
# define name for output csv file
custom_csv_filepath = "tracking_output_custom.csv"
# open the csv file in write mode
with open(custom_csv_filepath, mode="w", newline="") as file:
writer = csv.writer(file)
# write the header
writer.writerow(["frame", "ID", "x", "y", "width", "height"])
# write the data
for individual in ds_ff.individuals.data:
for frame in ds_ff.time.data:
x, y = ds_ff.position.sel(time=frame, individuals=individual).data
width, height = ds_ff.shape.sel(
time=frame, individuals=individual
).data
writer.writerow([frame, individual, x, y, width, height])
Clean-up#
To remove the output files we have just created, we can run the following code.
Total running time of the script: (0 minutes 3.481 seconds)