Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Compute distances and angles to regions of interest#
Compute distances, approach vectors, and both egocentric and allocentric boundary angles relative to regions of interest (RoIs).
Imports#
# For interactive plots: install ipympl with `pip install ipympl` and uncomment
# the following lines in your notebook
# %matplotlib widget
import matplotlib.pyplot as plt
import numpy as np
import xarray as xr
from movement import sample_data
from movement.kinematics import compute_velocity
from movement.plots import plot_centroid_trajectory
from movement.roi import PolygonOfInterest
Load sample dataset#
In this example, we will use the SLEAP_three-mice_Aeon_proofread example
dataset collected using the Aeon platform.
We only need the position data array, so we store it in a
separate variable.
ds = sample_data.fetch_dataset("SLEAP_three-mice_Aeon_proofread.analysis.h5")
positions: xr.DataArray = ds.position
Below we plot the image of the Aeon habitat within which the dataset was recorded.
habitat_fig, habitat_ax = plt.subplots(1, 1)
# Overlay an image of the habitat
habitat_image = ds.frame_path
habitat_ax.imshow(plt.imread(habitat_image))
habitat_ax.set_xlabel("x (pixels)")
habitat_ax.set_ylabel("y (pixels)")
habitat_fig.show()
The habitat is divided up into three main sub-regions. The cuboidal structure
on the right-hand-side of the habitat is the nest of the three individuals
taking part in the experiment. The majority of the habitat is an open
octadecagonal (18-sided) shape, which is the bright central region that
encompasses most of the image. This central region is surrounded by a
(comparatively thin) “ring”, which links the central region to the nest.
In this example, we will look at how we can use the functionality for regions
of interest (RoIs) provided by movement to analyse our sample dataset.
Define regions of interest#
In order to ask questions about the behaviour of our individuals with respect
to the habitat, we first need to define the RoIs to represent the separate
pieces of the habitat programmatically. Since each part of the habitat is
two-dimensional, we will use movement.roi.PolygonOfInterest
to describe each of them.
In the future, the movement plugin for napari will support creating regions of interest by clicking points and drawing shapes in the napari GUI. For the time being, we can still define our RoIs by specifying the points that make up the interior and exterior boundaries. So first, let’s define the boundary vertices of our various regions.
# The centre of the habitat is located roughly here
centre = np.array([712.5, 541])
# The "width" (distance between the inner and outer octadecagonal rings) is 40
# pixels wide
ring_width = 40.0
# The distance between opposite vertices of the outer ring is 1090 pixels
ring_extent = 1090.0
# Create the vertices of a "unit" octadecagon, centred on (0,0)
n_pts = 18
unit_shape = np.array(
[
np.exp((np.pi / 2.0 + (2.0 * i * np.pi) / n_pts) * 1.0j)
for i in range(n_pts)
],
dtype=complex,
)
# Then stretch and translate the reference to match the habitat
ring_outer_boundary = ring_extent / 2.0 * unit_shape.copy()
ring_outer_boundary = (
np.array([ring_outer_boundary.real, ring_outer_boundary.imag]).transpose()
+ centre
)
core_boundary = (ring_extent - ring_width) / 2.0 * unit_shape.copy()
core_boundary = (
np.array([core_boundary.real, core_boundary.imag]).transpose() + centre
)
nest_corners = ((1245, 585), (1245, 475), (1330, 480), (1330, 580))
Our central region is a solid shape without any interior holes. To create the appropriate RoI, we just pass the coordinates in either clockwise or counter-clockwise order.
central_region = PolygonOfInterest(core_boundary, name="Central region")
Likewise, the nest is also just a solid shape without any holes. Note that we are only registering the “floor” of the nest here.
nest_region = PolygonOfInterest(nest_corners, name="Nest region")
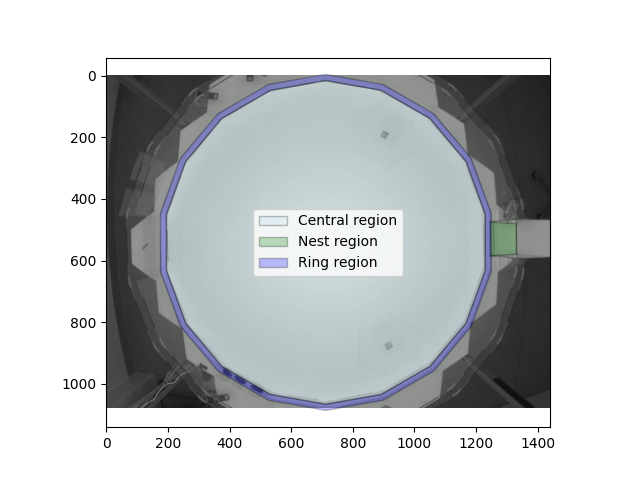
To create an RoI representing the ring region, we need to provide an interior
boundary so that movement knows our ring region has a “hole”.
PolygonOfInterest
can actually support multiple (non-overlapping) holes, which is why the
holes argument takes a list.
ring_region = PolygonOfInterest(
ring_outer_boundary, holes=[core_boundary], name="Ring region"
)
habitat_fig, habitat_ax = plt.subplots(1, 1)
# Overlay an image of the habitat
habitat_ax.imshow(plt.imread(habitat_image))
central_region.plot(habitat_ax, facecolor="lightblue", alpha=0.25)
nest_region.plot(habitat_ax, facecolor="green", alpha=0.25)
ring_region.plot(habitat_ax, facecolor="blue", alpha=0.25)
habitat_ax.legend()
habitat_fig.show()
View individual paths inside the habitat#
We can now overlay the paths that the individuals followed on top of our image of the habitat and the RoIs that we have defined.
habitat_fig, habitat_ax = plt.subplots(1, 1)
# Overlay an image of the habitat
habitat_ax.imshow(plt.imread(habitat_image))
central_region.plot(habitat_ax, facecolor="lightblue", alpha=0.25)
nest_region.plot(habitat_ax, facecolor="green", alpha=0.25)
ring_region.plot(habitat_ax, facecolor="blue", alpha=0.25)
# Plot trajectories of the individuals
mouse_names_and_colours = list(
zip(positions.individuals.values, ["r", "g", "b"], strict=False)
)
for mouse_name, col in mouse_names_and_colours:
plot_centroid_trajectory(
positions,
individual=mouse_name,
ax=habitat_ax,
linestyle="-",
marker=".",
s=1,
c=col,
label=mouse_name,
)
habitat_ax.set_title("Individual trajectories within the habitat")
habitat_ax.legend()
habitat_fig.show()
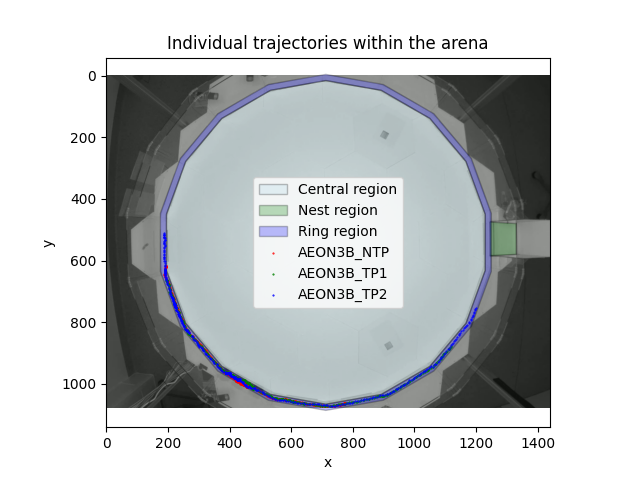
At a glance, it looks like all the individuals remained inside the
ring-region for the duration of the experiment. We can verify this
programmatically, by asking whether the ring_region
contained the individuals’ locations, at all recorded time-points.
# This is a DataArray with dimensions: time, keypoint, and individual.
# The values of the DataArray are True/False values, indicating if at the given
# time, the keypoint of individual was inside ring_region.
individual_was_inside = ring_region.contains_point(positions)
all_individuals_in_ring_at_all_times = individual_was_inside.all()
if all_individuals_in_ring_at_all_times:
print(
"All recorded positions, at all times,\n"
"and for all individuals, were inside ring_region."
)
else:
print("At least one position was recorded outside the ring_region.")
All recorded positions, at all times,
and for all individuals, were inside ring_region.
Compute the distance to the nest#
Defining RoIs means that we can efficiently extract information from our data
that depends on the location or relative position of an individual to an RoI.
For example, we might be interested in how the distance between an
individual and the nest region changes over the course of the experiment. We
can query the nest_region that we created for this information.
# Compute all distances to the nest; for all times, keypoints, and
# individuals.
distances_to_nest = nest_region.compute_distance_to(positions)
distances_fig, distances_ax = plt.subplots(1, 1)
for mouse_name, col in mouse_names_and_colours:
distances_ax.plot(
distances_to_nest.sel(individuals=mouse_name),
c=col,
label=mouse_name,
)
distances_ax.legend()
distances_ax.set_xlabel("Time (frames)")
distances_ax.set_ylabel("Distance to nest_region (pixels)")
distances_fig.show()
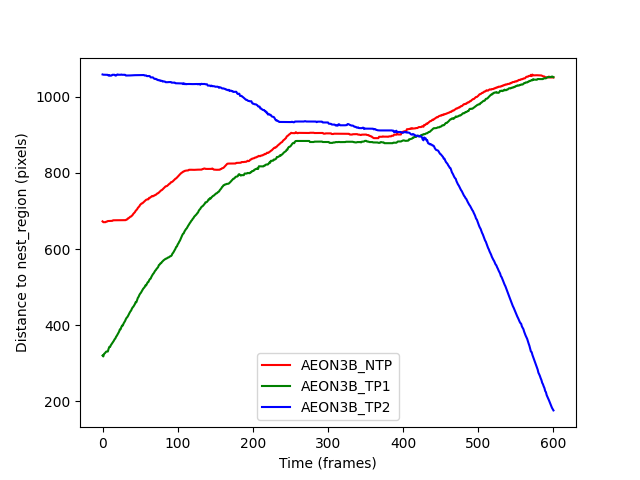
We can see that the AEON38_TP2 individual appears to be moving towards
the nest during the experiment, whilst the other two individuals are
moving away from the nest. The “plateau” in the figure between frames 200-400
is when the individuals meet in the ring_region, and remain largely
stationary in a group until they can pass each other.
One other thing to note is that
PolygonOfInterest.compute_distance_to()
is returning the distance “as the crow flies” to the nest_region.
This means that structures potentially in the way
(such as the ring_region walls) are not accounted for in this distance
calculation. Further to this, the “distance to a RoI” should always be
understood as “the distance from a point to the closest point within an RoI”.
If we wanted to check the direction of closest approach to a region, referred
to as the approach vector, we can use
PolygonOfInterest.compute_approach_vector().
The distances that we computed via
PolygonOfInterest.compute_distance_to() are just the
magnitudes of the approach vectors.
The boundary_only keyword#
From our plot of the distances to the nest, we saw a time-window
in which the individuals are grouped up, possibly trying to pass each other
as they approach from different directions.
We might be interested in whether they move to opposite walls of the ring
while doing so. To examine this, we can plot the distance between each
individual and the ring_region, using the same commands as above.
However, we get something slightly unexpected:
distances_to_ring_wall = ring_region.compute_distance_to(positions)
distances_fig, distances_ax = plt.subplots(1, 1)
for mouse_name, col in mouse_names_and_colours:
distances_ax.plot(
distances_to_ring_wall.sel(individuals=mouse_name),
c=col,
label=mouse_name,
)
distances_ax.legend()
distances_ax.set_xlabel("Time (frames)")
distances_ax.set_ylabel("Distance to closest ring_region wall (pixels)")
print(
"distances_to_ring_wall are all zero:",
np.allclose(distances_to_ring_wall, 0.0),
)
distances_fig.show()
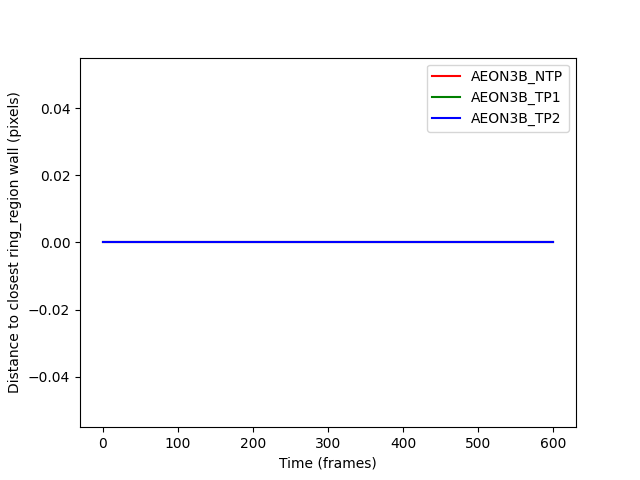
distances_to_ring_wall are all zero: True
The distances are all zero because when a point is inside a region, the closest point to it is itself.
To find distances to the ring’s walls instead, we can use
boundary_only=True, which tells movement to only look at points on
the boundary of the region, not inside it.
Note that for 1D regions (movement.roi.LineOfInterest),
the “boundary” is just the endpoints of the line.
Let’s try again with boundary_only=True:
distances_to_ring_wall = ring_region.compute_distance_to(
positions, boundary_only=True
)
distances_fig, distances_ax = plt.subplots(1, 1)
for mouse_name, col in mouse_names_and_colours:
distances_ax.plot(
distances_to_ring_wall.sel(individuals=mouse_name),
c=col,
label=mouse_name,
)
distances_ax.legend()
distances_ax.set_xlabel("Time (frames)")
distances_ax.set_ylabel("Distance to closest ring_region wall (pixels)")
print(
"distances_to_ring_wall are all zero:",
np.allclose(distances_to_ring_wall, 0.0),
)
distances_fig.show()
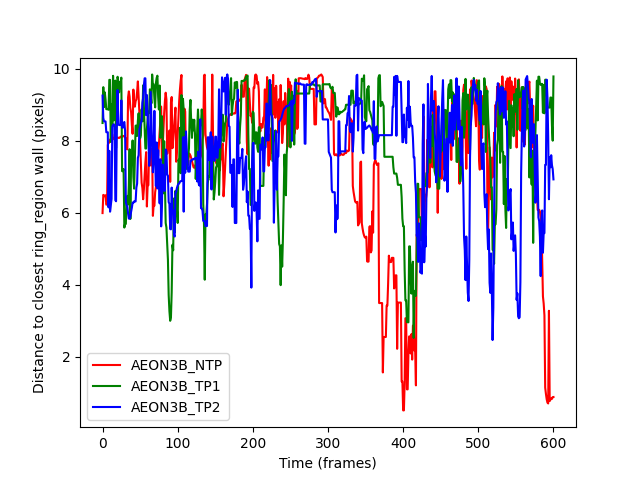
distances_to_ring_wall are all zero: False
The resulting plot looks much more like what we expect, but is again
not very helpful; we get the distance to the closest point on either
the interior or exterior wall of the ring_region. This means that we
can’t tell if the individuals do move to opposite walls when passing each
other. Instead, let’s ask for the distance to just the exterior wall.
# Note that the exterior_boundary of the ring_region is a 1D RoI (a series of
# connected line segments). As such, boundary_only needs to be False.
distances_to_exterior = ring_region.exterior_boundary.compute_distance_to(
positions
)
distances_exterior_fig, distances_exterior_ax = plt.subplots(1, 1)
for mouse_name, col in mouse_names_and_colours:
distances_exterior_ax.plot(
distances_to_exterior.sel(individuals=mouse_name),
c=col,
label=mouse_name,
)
distances_exterior_ax.legend()
distances_exterior_ax.set_xlabel("Time (frames)")
distances_exterior_ax.set_ylabel("Distance to exterior wall (pixels)")
distances_exterior_fig.show()
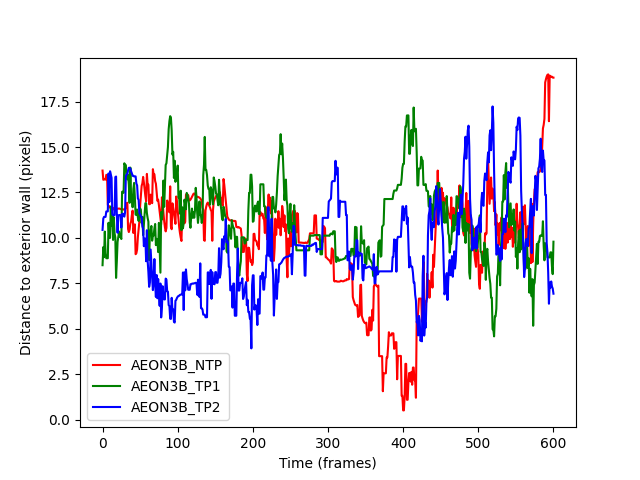
This output is much more helpful. We see that the individuals are largely the same distance from the exterior wall during frames 250-350, and then notice that;
Individual
AEON_TP1moves far away from the exterior wall,AEON3B_NTPmoves almost up to the exterior wall,and
AEON3B_TP2seems to remain between the other two individuals.
After frame 400, the individuals appear to go back to chaotic distances from
the exterior wall again, which is consistent with them having passed each
other in the ring_region and once again having the entire width of the
ring to explore.
Boundary angles#
Having observed the individuals’ behaviour as they pass one another in the
ring_region, we can begin to ask questions about their orientation with
respect to the nest. movement currently supports the computation of two
such “boundary angles”;
The allocentric boundary angle. Given a region of interest \(R\), reference vector \(\vec{r}\) (such as global north), and a position \(p\) in space (e.g. the position of an individual), the allocentric boundary angle is the signed angle between the approach vector from \(p\) to \(R\), and \(\vec{r}\).
The egocentric boundary angle. Given a region of interest \(R\), a forward vector \(\vec{f}\) (e.g. the direction of travel of an individual), and the point of origin of \(\vec{f}\) denoted \(p\) (e.g. the individual’s current position), the egocentric boundary angle is the signed angle between the approach vector from \(p\) to \(R\), and \(\vec{f}\).
Note that egocentric angles are generally computed with changing frames of reference in mind - the forward vector may be varying in time as the individual moves around the habitat. By contrast, allocentric angles are always computed with respect to some fixed reference frame.
For the purposes of our example, we will define our “forward vector” as the
velocity vector between successive time-points, for each individual - we can
compute this from positions using
movement.kinematics.compute_velocity().
We will also define our reference frame, or “global north” direction, to be
the direction of the positive x-axis.
forward_vector = compute_velocity(positions)
global_north = np.array([1.0, 0.0])
allocentric_angles = nest_region.compute_allocentric_angle_to_nearest_point(
positions,
reference_vector=global_north,
in_degrees=True,
)
egocentric_angles = nest_region.compute_egocentric_angle_to_nearest_point(
forward_vector,
positions[:-1],
in_degrees=True,
)
angle_plot, angle_ax = plt.subplots(2, 1, sharex=True)
allo_ax, ego_ax = angle_ax
for mouse_name, col in mouse_names_and_colours:
allo_ax.plot(
allocentric_angles.sel(individuals=mouse_name),
c=col,
label=mouse_name,
)
ego_ax.plot(
egocentric_angles.sel(individuals=mouse_name),
c=col,
label=mouse_name,
)
ego_ax.set_xlabel("Time (frames)")
ego_ax.set_ylabel("Egocentric angle (degrees)")
allo_ax.set_ylabel("Allocentric angle (degrees)")
allo_ax.legend()
angle_plot.tight_layout()
angle_plot.show()
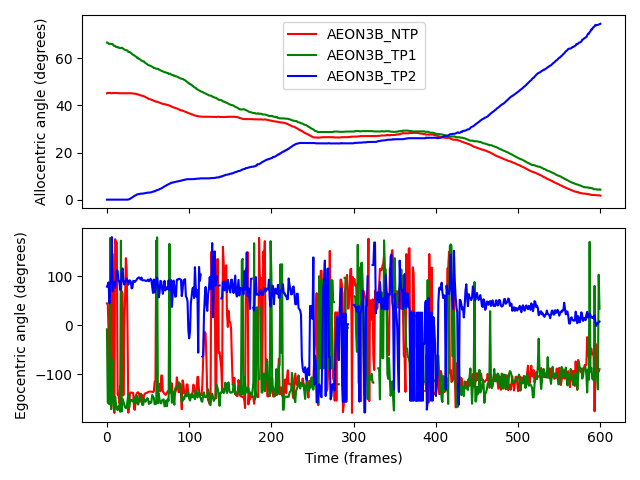
Allocentric angles show step-like behaviour because they only depend on an individual’s position relative to the RoI, not their forward vector. This makes the allocentric angle graph resemble the distance-to-nest graph (inverted on the y-axis), with a “plateau” during frames 200-400 when the individuals are (largely) stationary while passing each other.
Egocentric angles, on the other hand, fluctuate more due to their sensitivity to changes in the forward vector. Outside frames 200-400, we see trends:
AEON3B_TP2moves counter-clockwise around the ring, so its egocentric angle decreases ever so slightly with time - almost hitting an angle of 0 degrees as it moves along the direction of closest approach after passing the other individuals.The other two individuals move clockwise, so their angles show a gradual increase with time. Because the two individuals occasionally get in each others’ way, we see frequent “spikes” in their egocentric angles as their forward vectors rapidly change.
During frames 200-400, rapid changes in direction cause large fluctuations in the egocentric angles of all the individuals, reflecting the individuals’ attempts to avoid colliding with (and to make space to move passed) each other.
Total running time of the script: (0 minutes 1.333 seconds)