Note
Go to the end to download the full example code. or to run this example in your browser via Binder
Pupil tracking#
Look at eye movements and pupil diameter.
Imports#
import sleap_io as sio
import xarray as xr
from matplotlib import pyplot as plt
import movement.kinematics as kin
from movement import sample_data
from movement.filtering import rolling_filter
from movement.plots import plot_centroid_trajectory
Load the data#
We will use two datasets from the sample data module. These datasets involve
recordings of the eyes of mice placed on a rotating platform with different
visual stimuli. The uniform condition features a uniformly lit surround
stimulus, whereas the black condition was acquired in the dark. These
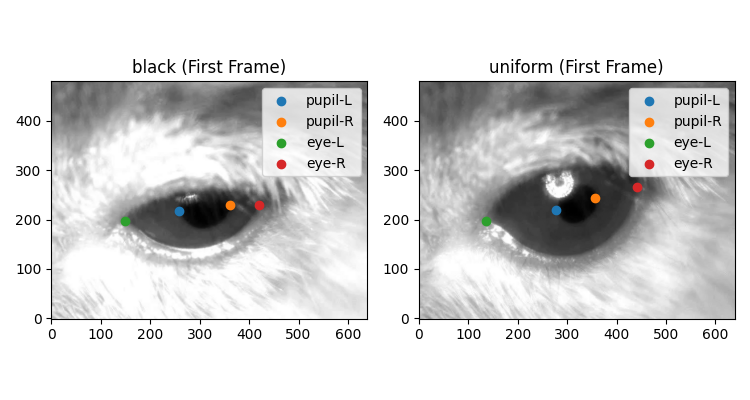
datasets were tracked using DeepLabCut (DLC) and include four keypoints:
two on either side of the pupil (pupil-L and pupil-R) and two on
either side of the eye (eye-L and eye-R).
ds_black = sample_data.fetch_dataset(
"DLC_rotating-mouse_eye-tracking_stim-black.predictions.h5",
with_video=True,
)
ds_uniform = sample_data.fetch_dataset(
"DLC_rotating-mouse_eye-tracking_stim-uniform.predictions.h5",
with_video=True,
)
# Save data in a dictionary.
ds_dict = {"black": ds_black, "uniform": ds_uniform}
Print the content of one of the datasets.
print(ds_dict["black"])
<xarray.Dataset> Size: 728kB
Dimensions: (time: 7000, space: 2, keypoints: 4, individuals: 1)
Coordinates:
* time (time) float64 56kB 0.0 0.025 0.05 0.075 ... 174.9 174.9 175.0
* space (space) <U1 8B 'x' 'y'
* keypoints (keypoints) <U7 112B 'pupil-L' 'pupil-R' 'eye-L' 'eye-R'
* individuals (individuals) <U12 48B 'individual_0'
Data variables:
position (time, space, keypoints, individuals) float64 448kB 258.8 .....
confidence (time, keypoints, individuals) float64 224kB 1.0 1.0 ... 1.0
Attributes:
source_software: DeepLabCut
ds_type: poses
fps: 40.0
time_unit: seconds
source_file: /home/runner/.movement/data/poses/DLC_rotating-mouse_ey...
frame_path: /home/runner/.movement/data/frames/rotating-mouse_eye-t...
video_path: /home/runner/.movement/data/videos/rotating-mouse_eye-t...
Explore the accompanying videos#
for ds_name, ds in ds_dict.items():
video = sio.load_video(ds.video_path)
# To avoid having to reload the video again we add it to ds as attribute
ds_dict[ds_name] = ds.assign_attrs({"video": video})
n_frames, height, width, channels = video.shape
print(f"Dataset: {ds_name}")
print(f"Number of frames: {n_frames}")
print(f"Frame size: {width}x{height}")
print(f"Number of channels: {channels}\n")
Dataset: black
Number of frames: 7000
Frame size: 640x480
Number of channels: 1
Dataset: uniform
Number of frames: 7000
Frame size: 640x480
Number of channels: 1
Plot first frame with keypoints#
fig, ax = plt.subplots(1, 2, figsize=(7.5, 4))
for i, (da_name, ds) in enumerate(ds_dict.items()):
ax[i].imshow(ds.video[0], cmap="gray") # plot first video frame
for keypoint in ds.keypoints.values:
x = ds.position.sel(time=0, space="x", keypoints=keypoint)
y = ds.position.sel(time=0, space="y", keypoints=keypoint)
ax[i].scatter(x, y, label=keypoint) # plot keypoints
ax[i].legend()
ax[i].set_title(f"{da_name} (First Frame)")
ax[i].invert_yaxis() # because the dataset was collected flipped
plt.tight_layout()
plt.show()
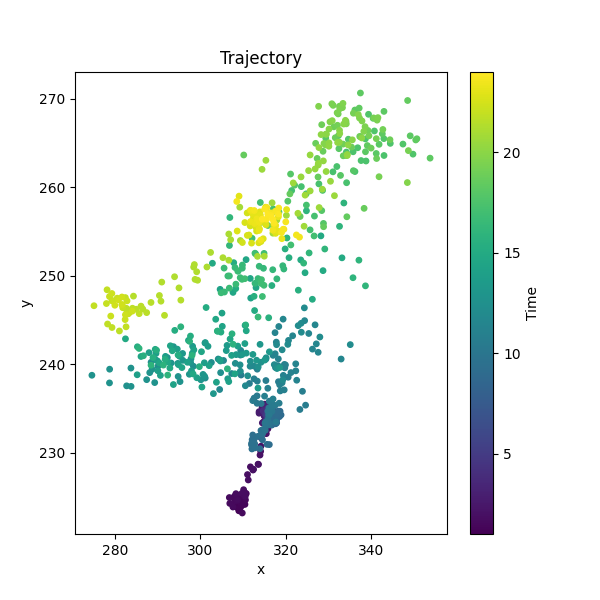
Pupil trajectory#
A quick plot of the trajectory of the centre of the pupil using
movement.plots.plot_centroid_trajectory().
time_window = slice(1, 24) # seconds
position_black = ds_black.position.sel(time=time_window) # data array to plot
fig, ax = plot_centroid_trajectory(
position_black, keypoints=["pupil-L", "pupil-R"]
)
fig.show()
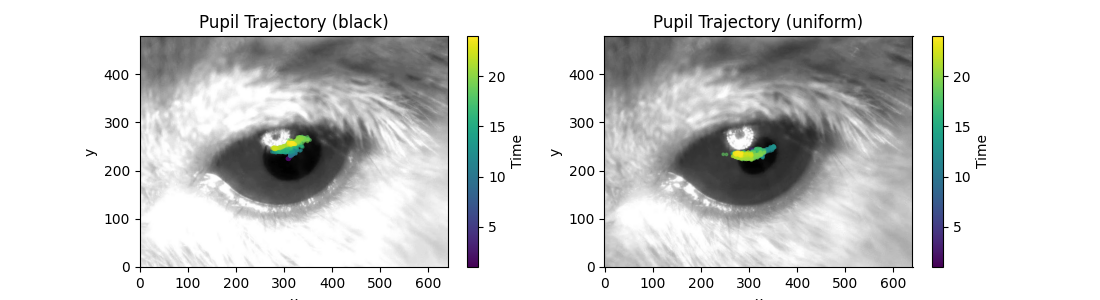
Pupil trajectories on top of video frame#
We can look at pupil trajectories plotted on top of a video frame.
fig, ax = plt.subplots(1, 2, figsize=(11, 3))
for i, (ds_name, ds) in enumerate(ds_dict.items()):
ax[i].imshow(ds.video[100], cmap="gray") # Plot frame 100 as background
plot_centroid_trajectory(
ds.position.sel(time=time_window), # Select time window
ax=ax[i],
keypoints=["pupil-L", "pupil-R"],
alpha=0.5,
s=3,
)
ax[i].invert_yaxis()
ax[i].set_title(f"Pupil Trajectory ({ds_name})")
fig.show()
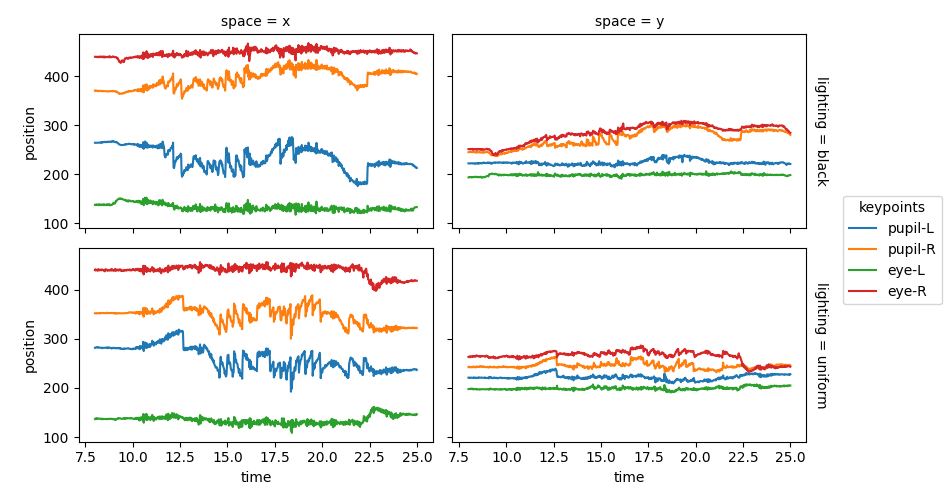
Keypoint positions over time#
For the rest of this example we are only interested in the position data.
For convenience, We will combine the two position arrays into a single
array with a new dimension called lighting.
positions = xr.concat([ds_black.position, ds_uniform.position], "lighting")
positions.coords["lighting"] = ["black", "uniform"]
Define plotting parameters for reuse.
plot_params = {
"x": "time",
"hue": "keypoints",
"col": "space",
"row": "lighting",
"aspect": 1.5,
"size": 2.5,
}
sel = {"time": slice(8, 25)}
Plot the keypoint positions over time.
positions.sel(**sel).squeeze().plot.line(**plot_params)
plt.subplots_adjust(right=0.85) # Make space on the right for the legend
plt.show()
Normalised keypoint positions over time#
Normalising the pupil’s position relative to the midpoint of the eye reduces the impact of head movements or artefacts caused by camera movement. By subtracting the position of the eye’s midpoint, we effectively transform the data into a moving coordinate system, with the eye’s midpoint as the origin. In the rest of the example, the normalised data will be used.
eye_midpoint = positions.sel(keypoints=["eye-L", "eye-R"]).mean("keypoints")
positions_norm = positions - eye_midpoint
We plot the x and y positions again, but now using the normalised data.
positions_norm.sel(**sel).squeeze().plot.line(**plot_params)
plt.subplots_adjust(right=0.85)
plt.show()
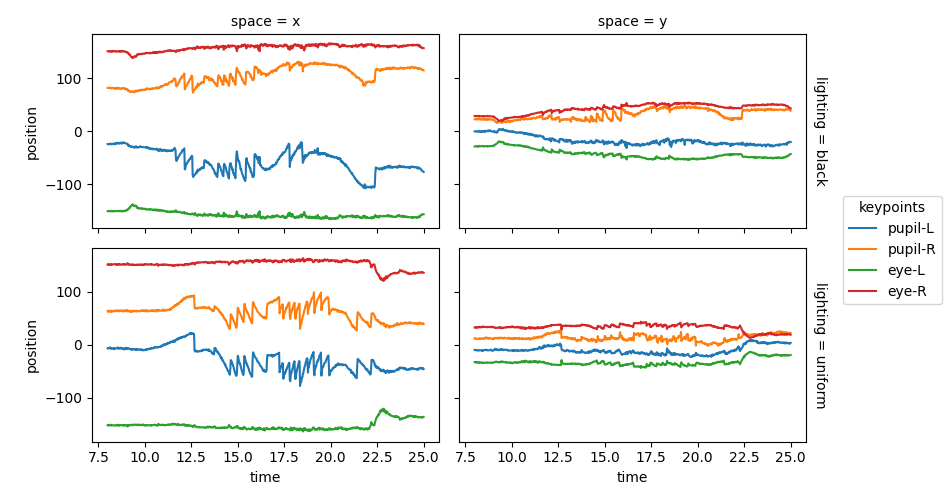
Pupil position over time#
To look at pupil position—and later also velocity—over time, we use the
pupil centroid (in this case the midpoint between keypoints pupil-L and
pupil-R). The keypoint pupil-C is assigned using
xarray.DataArray.assign_coords().
pupil_centroid = (
positions_norm.sel(keypoints=["pupil-L", "pupil-R"])
.mean("keypoints")
.assign_coords({"keypoints": "pupil-C"})
)
The pupil centroid keypoint pupil-C is be added to the positions_norm
using xarray.concat().
positions_norm = xr.concat([positions_norm, pupil_centroid], dim="keypoints")
Now the position of the pupil centroid pupil-C can be plotted.
positions_norm.sel(keypoints="pupil-C", **sel).squeeze().plot.line(
x="time", hue="space", row="lighting", aspect=3.5, size=1.5
)
plt.show()
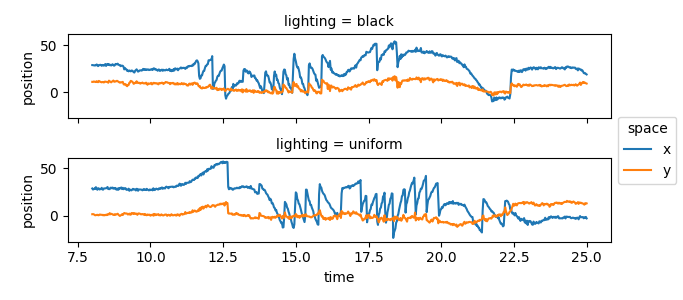
Pupil velocity over time#
In these experiments, the mouse is being rotated clock- or anti-clock-wise, triggering the vestibulo-ocular reflex. This reflex involves the vestibular system in the inner ear, that detects head motion and adjusts eye position to maintain stable vision.
When the head turns beyond the range that the vestibulo-ocular reflex
can compensate for, a quick, ballistic eye movement is triggered to
shift gaze to a new fixation point. These fast eye movements are seen in the
previous plot but become even more obvious when the velocity of the pupil
centroid is plotted. To do this, we use
movement.kinematics.compute_velocity()
to calculate the velocity of the eye movements.
pupil_velocity = kin.compute_velocity(positions_norm.sel(keypoints="pupil-C"))
pupil_velocity.name = "pupil velocity"
pupil_velocity.sel(**sel).squeeze().plot.line(
x="time", hue="space", row="lighting", aspect=3.5, size=1.5
)
plt.show()
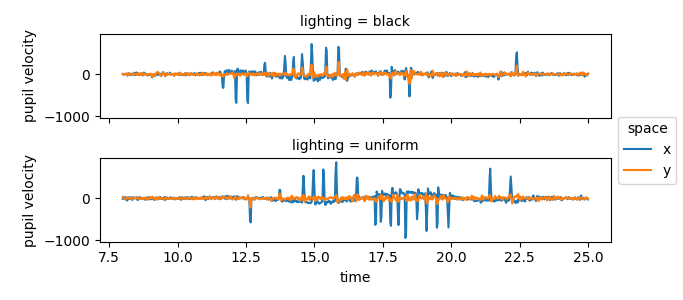
The positive peaks correspond to rapid eye movements to the right, the negative peaks correspond to rapid eye movements to the left.
Pupil diameter#
Here we define the pupil diameter as the distance between the two pupil
keypoints. We use movement.kinematics.compute_pairwise_distances()
to calculate the Euclidean distance between pupil-L and pupil-R.
pupil_diameter: xr.DataArray = kin.compute_pairwise_distances(
positions_norm, dim="keypoints", pairs={"pupil-L": "pupil-R"}
)
pupil_diameter.name = "pupil diameter"
Now the pupil diameter can be plotted.
pupil_diameter.plot.line(x="time", hue="lighting")
plt.show()
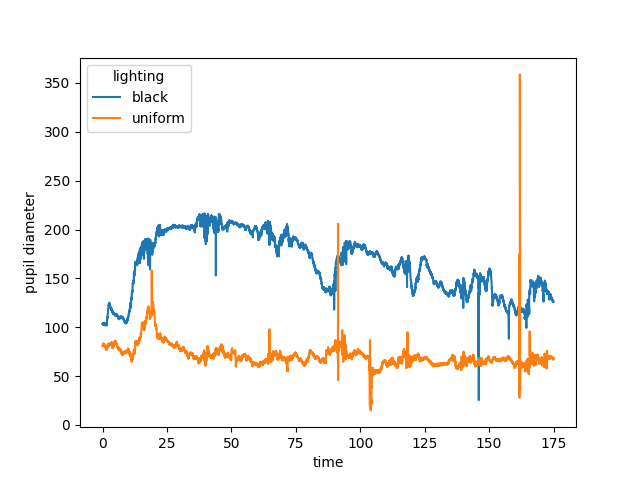
The plot of the pupil diameter looks noisy. The very steep peaks are unlikely to represent real changes in the pupil size. In fact, these steep peaks are probably caused by tracking errors during blinking or squinting. By looking at the distance between the two eye keypoints we can get an idea of whether (and when) the animal is blinking or squinting.
distance_between_eye_keypoints: xr.DataArray = kin.compute_pairwise_distances(
positions_norm, dim="keypoints", pairs={"eye-L": "eye-R"}
)
distance_between_eye_keypoints.name = "distance (eye-L - eye-R)"
# Combine the datasets into one DataArray
combined = xr.concat(
[distance_between_eye_keypoints, pupil_diameter], dim="variable"
)
combined = combined.assign_coords(
variable=["distance (eye-L - eye-R)", "pupil diameter"]
)
# Plot the distance between the eye keypoints alongside the pupil diameter
combined.plot.line(
x="time", row="lighting", hue="variable", figsize=(8, 4), add_legend=False
)
labels = combined.coords["variable"].values
plt.legend(labels, loc="center", bbox_to_anchor=(0.5, 1.4), ncol=2)
plt.xlabel("time (s)")
[ax.set_ylabel("distance (pixels)") for ax in plt.gcf().axes]
plt.show()
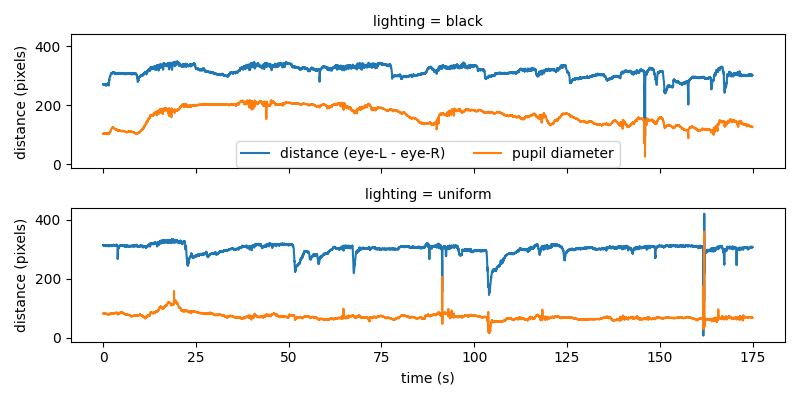
We indeed see that the sharp peaks in pupil diameter correspond to abrupt changes of distance between the two eye keypoints. Compared to fast eye movements and blinking, changes in pupil size are slow. Filters can be applied to reduce noise and make underlying trends in pupil diameter clearer.
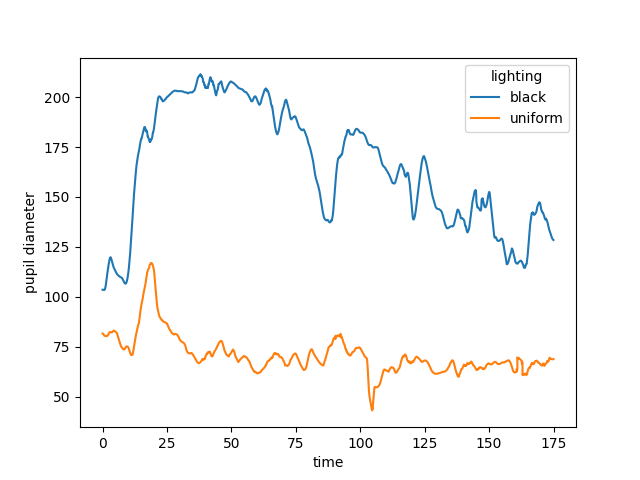
Smooth pupil diameter#
A rolling mean (moving average) filter is used here to smooth the data by
averaging a specified number of data points (window_len).
We achieve this by calling movement.filtering.rolling_filter()
with the statistic="mean" option.
window_len = 80
mean_filter = rolling_filter(
pupil_diameter, window=window_len, statistic="mean"
)
Now the filtered pupil diameter can be plotted.
mean_filter.plot.line(x="time", hue="lighting")
plt.show()
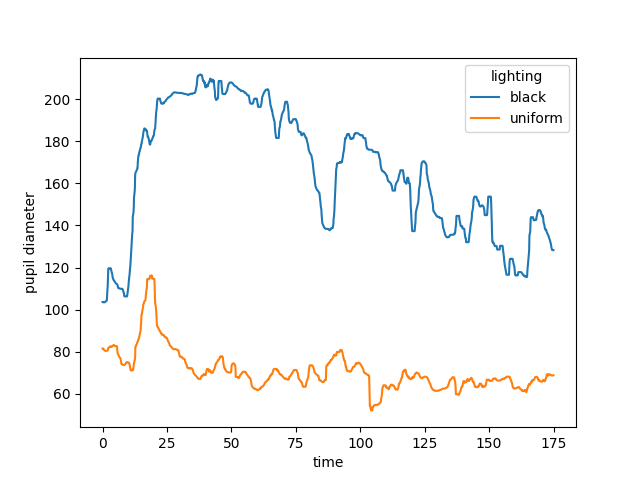
Instead of the mean, we could also use the median, which is the default
option for the statistic argument, and should be more robust to outliers.
mdn_filter = rolling_filter(pupil_diameter, window_len, statistic="median")
mdn_filter.plot.line(x="time", hue="lighting")
plt.show()
See also
Smooth pose tracks example.
Total running time of the script: (0 minutes 3.299 seconds)